Open Computational Research Study - A Proof of Concept
08 Mar 2018
Computational research is often published without the code and computational environment that generated the results, failing to meet openness and reproducibility criteria that are demanded by a growing number of researchers and journals1, 2, 3.
Here I'm presenting a proof of concept, how code and results of a complex computational research project can be published in a way to satisfy important openness and reproducibility requirements. Requiring only a Docker installation, it should be possible for users on any system to comprehend and reproduce any and all parts of a project shared this way.
Specifically, the concept presented here addresses two issues not typically considered when publishing scientific code:
- Research code published without its computational dependencies may become exceedingly difficult to compile and execute, to the point that a rewrite of the code from the ground up becomes a preferred solution over trying to recreate the computational environment4, 5.
- A large computational project that is not sufficiently documented (which computations, when, why and how?) might be reproducible in the sense that a single run command creates all the figures shown the paper, but it might not be comprehensible, failing to show which parts of the code affect which results. Moreover, if the simulations are computationally expensive, it might be of importance to be able to identify and reproduce only selected parts of the project.
Both points may be equally detrimental to any effort of straightforwardly reproducing the computational results of a study. To demonstrate the concept I'm proposing here, I have created an example study that anyone having a Docker installation should be able to reproduce and expand.
Below I'm describing this example study in detail and explain how it is implemented. I would love to hear your thoughts and feedback!
An Example Study
I've created a small computational project to demonstrate a way how the above issues may be solved in practice. My proposed solution relies on two main components: Docker, to provide an isolated computational environment that contains all needed dependencies and Sumatra, to provide a lab notebook of the computational project to be able to comprehend and replicate parts of a large and complex computational study.
The downloadable archive for this example project contains the Python source code and the data it generated. Additionally included is version control (.git/) and simulation record information (.smt/). Finally, I provided scripts to access the computational environment through Docker for Linux, Mac and Windows 7 systems (I'm not able to test on other Windows versions).
For Linux systems for example, access_lab_linux.sh contains
#!/bin/bash docker run -it -p 127.0.0.1:8015:8000 \ --user="$(id -u):$(id -g)" \ -v $(pwd):/home/lab \ felix11h/docker-open-comp-rsc \ /bin/bash -c \ 'cd /home/lab/comp/; screen -d -m smtweb --allips; source ../startup_messg_linux.sh; bash'
Running the script first tells Docker to run the image felix11h/open-com-rsc. As the image is very likely not found on your machine, the image will be automatically downloaded from Docker Hub. This is convenient for users, but has another advantage: As the image was built by Docker Hub, you can verify the contents of the image by inspecting the associated Dockerfile. In this case the image was built from the following Dockerfile
FROM ubuntu:16.04 MAINTAINER felix11h.dev@gmail.com USER root RUN apt-get -qy update RUN apt-get install -qy apt-utils python python-dev python-pip git screen RUN pip install --upgrade pip RUN pip install numpy scipy matplotlib sumatra gitpython configparser RUN useradd -ms /bin/bash docker USER docker
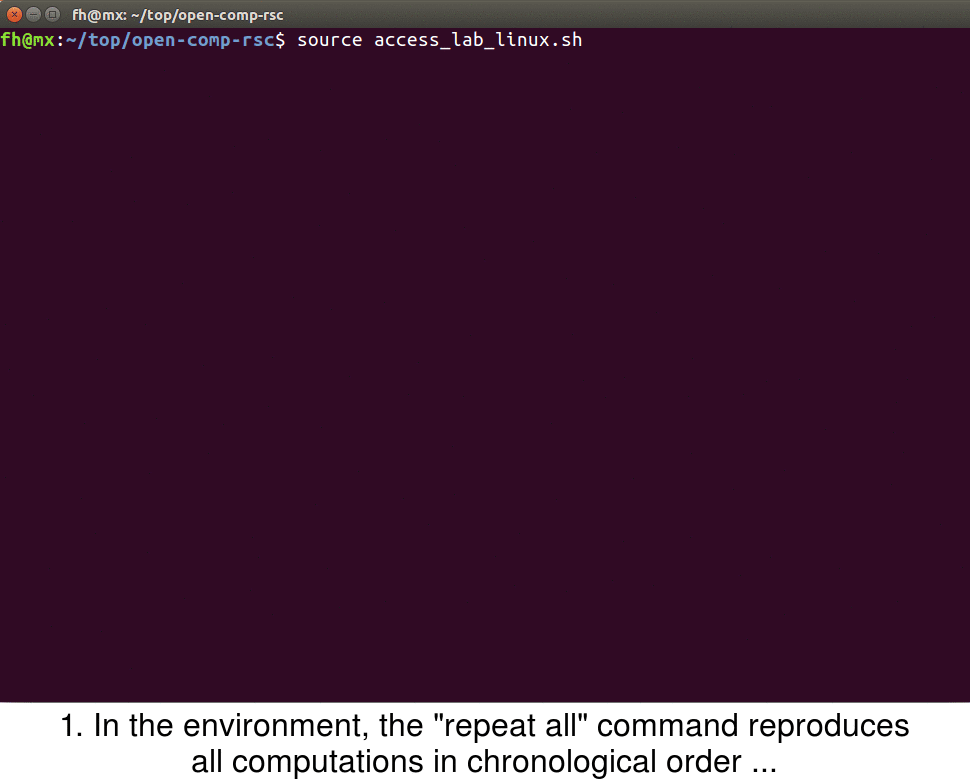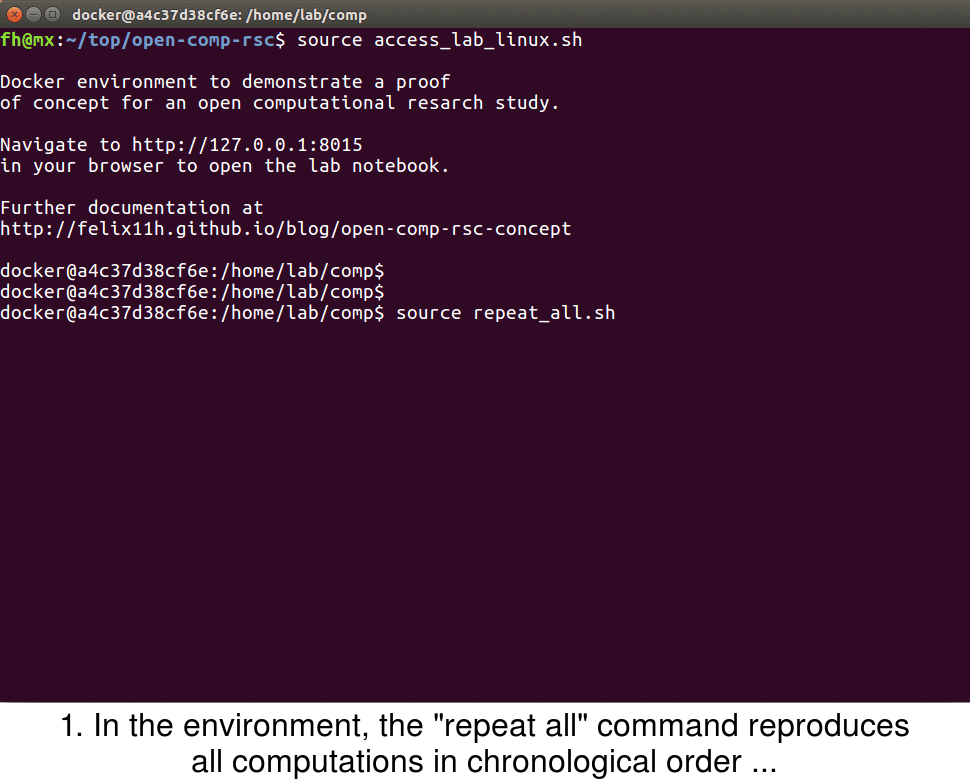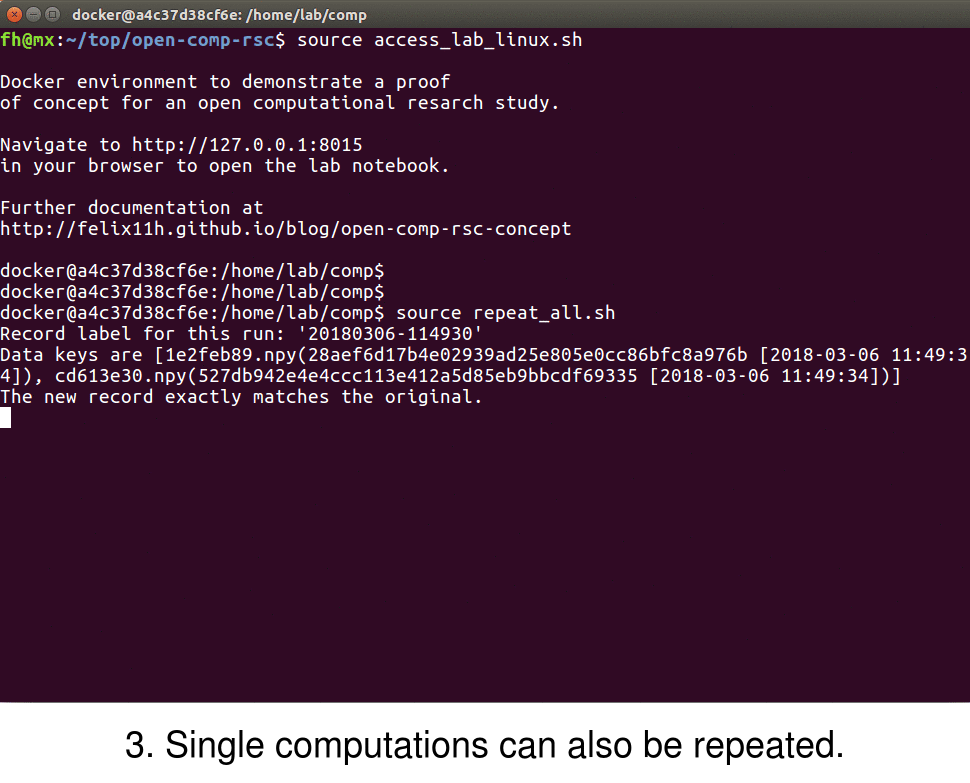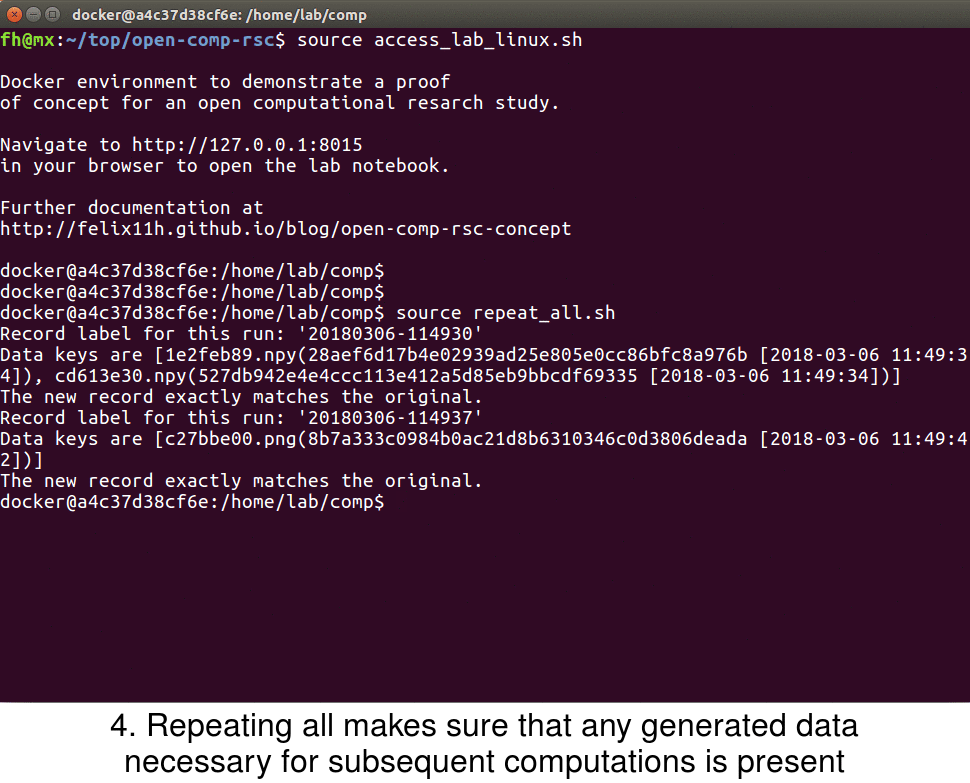
Once the image is downloaded, a few more things happen: The current directory containing code and data is mounted within the Docker container, the address 0.0.0.0:8000 in the container is mapped to 127.0.0.1:8015 on the host, the Sumatra web interface smtweb is run in a background screen and, finally, a welcome message is printed once the container is accessed.
The 5 steps from downloading the archive to accessing the container are illustrated in the animation below
Sumatra lab notebook
With the Docker container the full research environment is now accessible to the user. To access the lab notebook for example, one can simply navigate the browser to the address printed in the terminal. On Linux systems one would open http://127.0.0.1:8015.
This lab notebook was automatically generated by Sumatra while running the original simulations. The web interface lists all simulation records or, in the alternative view, all data of the computational project. The notebook contains the full information of the simulation: version of the code, parameters, input and output data information, dependencies, machine specification and standard output – the full provenance of the results of this simulation is available. The animation below gives an overview of the lab notebooks contents of the example study
Reproducibility
Sumatra also provides the ability to easily replicate and verify the results of a given simulation via the smt repeat command. To verify, for example, the results of a computation with label d6bcfd41, one can use the command
$ smt repeat d6bcfd41
This repeats the simulation d6bcfd41 with the exact parameters and specifications as the original computation and records the output of this process under a new label. The printed output should verify that the reproduced and original simulation match exactly. Note that this doesn't only compare to output data file but checks various parameters that might influence results
$ smt diff d6bcfd41 20180111-121253 Record 1 : d6bcfd41 Record 2 : 20180111-121253 Executable differs : no Code differs : no Repository differs : no Main file differs : no Version differs : no Non checked-in code : no Dependencies differ : no Launch mode differs : no Input data differ : no Script arguments differ : no Parameters differ : no Data differ : no
In the example study I provide the script repeat_all.sh to repeat all computations of the project in chronological order
#!/bin/bash smt list -r | xargs -L1 smt repeat
With this all computations are executed in their original order – the full study can be reproduced with this command. This also allows for sharing of the computational project without any generated data, which can be helpful if this data is large. The final animation shows the smt repeat command in the example study
Feedback?
Publishing computational research projects in this proposed format should provide direct access to the results and a straightfoward way to reproduce and interact with the code and generated results. With the example study I provided I wanted to show that such an implementation is possible, however the concept still needs to be proven on the scale of a full study.
As part of my participation in the Open Science Fellows Program, I'm planning to publish my computational neuroscience research following this format. For this, and this is part of my motivation for this post, I want to ask for your feedback. Does the concept I present make for an open, reproducible computational research study? What aspects are you missing? What problems are you foreseeing? I would be very grateful for your feedback!
Footnotes
Topalidou, M., Leblois, A., Boraud, T. & Rougier, N. P. A long journey into reproducible computational neuroscience. Front. Comput. Neurosci. 9, (2015).
Rougier, N. P. et al. Sustainable computational science: the ReScience initiative. arXiv:1707.04393 (2017)